Addressing the Concerns of a Critic and the Case for Intelligent Design
In 2008, Baker Books published my book, The Cell’s Design. Overall, the reaction to the book has been positive. But there have been a few reviews that were critical. One of note was written by microbiologist Frank Steiner. A recent discovery regarding the structure of an enzyme by researchers from Germany helps address the central point made by Steiner, and in doing so actually strengthens the case for the intelligent design of biochemical systems.
I don’t like to be criticized—not. one. bit. But if I am honest with myself, there is almost always some measure of validity to the critical comments directed my way.
As my friend, who is a pastor, once told me, “criticism is a gift.” If you are willing to address your identified shortcomings, you have the chance to become an even better version of yourself. This principle also applies to Christian apologetics: If you take criticisms seriously and adequately address them, your arguments for the Christian faith become that much more powerful.
Generally speaking, the reaction to my book The Cell’s Design has been positive. But there have been a few reviews that were less than stellar. Perhaps the most critical of all was a review written by microbiologist Frank Steiner for the Reports of the National Center for Science Education.
After careful reflection, I have come to conclude that many of the issues Steiner has with The Cell’s Design are unsubstantial and largely unfounded. Nevertheless, one point he raised has some merit. Fortunately, a recent discovery by researchers from Germany about the structure of the enzyme F1-F0-ATPase helps address Steiner’s point—and in doing so, actually strengthens my argument for the intelligent design of biochemical systems.
An Important Criticism
I am not going to respond to all of Steiner’s criticism. Instead, I will focus on the concern I think has the most validity, the one that Steiner refers to as “the principle scientific problem that I have with Rana’s approach.” Namely, “Each of his ‘design’ examples—or what he produces as evidence for design—is presented in such general terms that he has glossed over the diversity represented in related systems or organisms, all of which can be accounted for by evolutionary mechanisms.”
To illustrate, Steiner’s complaint, consider the supramolecular architecture of the enzyme F1-F0-ATPase.
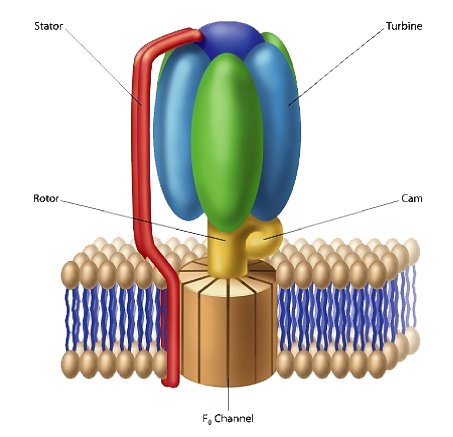
As I discussed in The Cell’s Design, F1-F0-ATPase, is a molecular-scale rotary motor, ubiquitous in the biological realm (see figure 1). This protein complex plays a central role in harvesting energy for cellular use. F1-F0-ATPase associates with cell membranes. The F1 portion of the complex is mushroom-shaped and extends above the membrane’s surface. The “button of the mushroom” literally corresponds to an engine turbine. The F1-F0-ATPase turbine interacts with the part of the complex that looks like a “mushroom stalk.” This stalk-like component functions as a rotor. The flow of positively charged hydrogen ions (or in some instances sodium ions) through the F0 component, embedded in the cell membrane, drives the rotation of the rotor. A rod-shaped protein structure that also extends above the membrane surface serves as a stator. This protein rod interacts with the turbine, holding it stationary as the rotor rotates.
The electrical current that flows through the channels of the F0 complex is transformed into mechanical energy, which then drives the rotor’s movement. A cam that extends at a right angle from the rotor’s surface causes displacements of the turbine. These back-and-forth motions are used to produce ATP (adenosine triphosphate). The cell uses this compound as a source of chemical energy to drive the operation of cellular processes.
Purposeful Processes
In The Cell’s Design and elsewhere, I argue that the discovery of enzymes with machine-like domains, such as F1-F0-ATPase, revitalize the Watchmaker argument. Popularized by William Paley in the eighteenth century, this argument states that as a watch requires a watchmaker, so too, life requires a Creator.
This simple, yet powerful, argument has been challenged by skeptics like David Hume who assert that the necessary conclusion of a Creator, based on analogical reasoning, is only compelling if there is a high degree of similarity between the objects that form the analogy. Skeptics have long argued that nature and a watch are sufficiently dissimilar so that the conclusion drawn from the Watchmaker argument is unsound.
Due to the striking similarity between the machine parts of these enzymes and man-made devices, the discovery of enzymes with domains that are direct analogs to man-made devices addresses this concern.
Now for Steiner’s point. The structure of F1-F0-ATPase shown in figure 1 is a generalized structure. Steiner correctly pointed out that the structure of this enzyme varies from organism to organism. For example, the F0 ring embedded in the membrane is comprised of several copies of the same protein (called the c protein). The number of c protein subunits in the F0 ring is constant for a given species, but varies from species to species, with some organisms having as few as 10 copies and others as many as 15 copies.
Steiner would argue that this variability reflects the outworking of the evolutionary process, making no sense from a design standpoint. Why wouldn’t the Creator simply make a single version of the F1-F0-ATPase complex? Steiner’s assumption is that this structural variability has no functional purpose. It simply reflects the sloppy, randomness of the evolutionary process.
But, as it turns out, new work by scientists from Germany indicates that the variable number of c subunits has functional significance.1 The number of c subunits determines how much “electrical current” must flow through the F0 subunit before the F1 portion of the enzyme can make a molecule of ATP. These researchers note that “c-subunit amino acid sequences [appear to be] adapted to fit best the c-ring stoichiometry that sufficiently fulfills the bioenergetic requirements of each individual species.”2
Generalizing the implications of this study, it could very well be that much of the biochemical diversity observed throughout nature may actually have a functional basis. This realization has gone unrecognized because of the excessive influence of the evolutionary paradigm. In other words, the biochemical diversity may not be haphazard at all, but reflects the unique functional requirements of different organisms.
In his review, Steiner complained that I “start with the foregone conclusion that the cell is designed…Thereafter, everything is presented through this lens.” I could argue the same is true for adherents of the evolutionary paradigm. That is, they start with the foregone conclusion that evolution is a fact and then view everything through that lens. Looking through the glasses of evolution, biochemical diversity stems from the randomness of the evolutionary process.
It may well be that viewing biochemistry from an evolutionary perspective has hampered our understanding of the functional importance of biomolecular diversity. Instead of seeing biodiversity as having a rational basis—providing functional insight into the operation of biochemical systems—researchers unduly influenced by the evolutionary paradigm have simply dismissed it as the outworking of random processes.
Human designers often employ variations of a generalized design to adapt the generalized design to a specific set of demands. Why wouldn’t this be the case for the design of biochemical systems? To say it another way, could it be that “the diversity represented in related systems of organisms” reflects the intentional work of a Creator, adapting a generalized design for the specific set of demands confronting different organisms? If so, this possibility deepens the similarity between man-made designs and the structure and function of biochemical systems
Endnotes
- Denys Pogoryelov et al., “Engineering Rotor Ring Stoichiometries in the ATP Synthase,” Proceedings of the National Academy of Sciences, USA 109 (2012): E1599–E1608.
- Ibid.






