Splicing Together the Case for Design, Part 1 (of 2)
At 13 years old I left Little League and took the next step in my baseball “career”—I joined the Babe Ruth League. Soon after it became apparent I didn’t have a future as a major leaguer.
It wasn’t the major league dimensions of the field or leading off of the bases before the pitch passed the batter that caused me problems. It was some of the finer points of the game, like picking up signs from the third base coach.
Our third base coach would go through an elaborate series of signs (touching his hat, elbows, nose, etc.) to give instructions to the batter and base runners. Embedded somewhere in this sequence was the indicator signal (say, right hand across the chest) then the instruction (bunt). The extra signs were meant to confuse the other team so they couldn’t make sense of the message.
The only problem was that this complex system invariably confused me. It was so bad that when I came up to bat, the coach would call time out, have me walk over to him, then whisper instructions to me. It was humiliating to say the least. It was a great system, but just too complicated for me, and I was never able to fully grasp it.
In biochemistry the splicing code is a complex system of signals. Fortunately, unlike me as a thirteen-year-old ball player, a team of biochemists is making progress in understanding these biochemical signals.1 This is no small feat. The code, which plays a key role in the expression of genetic information, is extremely confusing and has been difficult for scientists to grasp.
This new insight will help scientists understand how gene expression is controlled. And it also reveals additional evidence for the intelligent design of biochemical systems.
In part 1 of this TNRTB series, I describe the splicing process and touch on how it evinces the work of a Creator. Next week, I will describe the efforts by the international team to decipher the splicing code and show how their insights help advance the case for biochemical intelligent design.
Gene Splicing
In eukaryotes genes consist of regions that specify the amino acid sequence of protein chains (called exons) interrupted by other regions that don’t code for anything (called introns).
After the gene is copied into a mRNA molecule, the intron sequences are excised and the exons spliced together by a protein-RNA complex known as a spliceosome. (The figure below illustrates this process.)
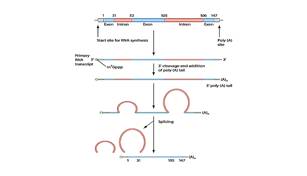
Splicing is an extremely precise process. Errors can cause major problems for organisms and are even responsible for some human diseases. The latter occurs when splicing errors fatally distort or destroy the information (temporarily stored in mRNA) needed to assemble protein chains at ribosomes. And proteins produced improperly cannot carry out their functional roles in the cell.
In Essentials of Molecular Biology, George Malacinski points out, “A cell cannot, of course, afford to miss any of the splice junctions by even a single nucleotide, because this could result in an interruption of the correct reading frame, leading to a truncated protein.”2
Alternate Splicing
In light of this constraint, it is remarkable that the spliceosome can splice the same mRNA in different ways in order to produce a range of functional proteins. This process (called alternate splicing) occurs because the spliceosome does not necessarily use all the splice sites, which allows the cell to produce several different proteins from the same mRNA, and ultimately from the same gene.
Proteins involved in the splicing process help determine the splicing pattern of mRNA. For example, some proteins bind to splice sites to block access by the spliceosome. By varying the binding pattern of these proteins, an assortment of mRNAs can be produced.
The cell also achieves alternate splicing through the use of different promoters associated with the same gene.3 Each promoter produces an mRNA spliced differently by the spliceosome complex.
Alternate splicing can also be mediated by varying the length of the mRNA’s poly (A) tail. This structural feature is generated after the mRNA molecule is produced. A protein called poly (A) polymerase adds about 200 adenine nucleotides to the last position of the mRNA molecule to form what biochemists call a poly (A) tail. This tail imparts stability to the mRNA molecule. Apparently the poly (A) tail also helps direct the spliceosome to specific splice junctions along the mRNA strand.
Alternate Gene Splicing and the Case for Intelligent Design
Structuring genes to have non-coding regions (introns) interspersed between coding regions (exons) is an elegant strategy. It allows a single gene to house simultaneously information for producing a range of proteins. In fact, nearly 95 percent of human genes undergo alternate splicing. This means that the 20,000 to 30,000 genes in the human genome actually specify more proteins (perhaps more than 100,000) than are directly encoded into the DNA.
Given the exactness of the splicing process and its sensitivity to errors, it’s astounding that alternate splicing occurs at all. For the cell to successfully carry out alternate splicing, the nucleotide sequences and the placement of exons have to be carefully orchestrated. And as I argue in The Cell’s Design, the elegance and careful design needed for this delicate process suggest that a Mind is behind it all.
But the alternate splicing of genes is just the indicator signal for design. As I will describe next week, the actual sign is the splicing code.
There are two key sites within the regulatory region of a gene: the promoter and the operator. The promoter serves as the binding site for a massive protein complex called RNA polymerase. This enzyme initiates gene expression by producing a messenger RNA molecule, which contains a copy of the information found in the protein coding region of the gene.
| Part 1 | Part 2 |
Endnotes
- Yoseph Barash et al., “Deciphering the Splicing Code,” Nature 465 (2010): 53–59.
- George M. Malacinski, Essentials of Molecular Biology, 4th ed. (Boston: Jones and Bartlett, 2003), 261–65.
- Gene structure is complex, largelyconsisting of two regions: the protein coding region and the regulatory region. The protein coding region contains the information the cell’s biochemical machinery needs to produce the protein chain encoded by that gene. The regulatory region controls the expression of the gene, and hence, the production of the protein. In effect, the regulatory region consists of “on/off switches” and “volume control knobs” that regulate gene expression.





